How to Plot a World Map Using Python and GeoPandas
Learn how to plot maps of the world, continents, and countries using Python and GeoPandas.
Over the past few weeks I’ve been exploring the International Disaster Database (EM-DAT), which provides data on over 25,000 mass disasters from the year 1900 to present day.
I first came across the database after discovering Juliana Negrini de Araujo’s fantastic Jupyter Notebook which provides some high-level exploration of the dataset.
In many ways, the EM-DAT dataset is my first foray into analyzing natural disasters with Python.
I’ve spent some time building on Juliana’s work, cleaning up some code, and providing some additional detail and commentary — today I’m excited to publish the article. I hope it helps the community, and at the very least, it serves as documentation for myself as I continue exploring the EM-DAT dataset.
Overall, the goal of this article is to provide a detailed introduction to the EM-DAT natural disaster dataset, serving as a starting point for anyone else in the community who wants to study natural disasters through a data science lens.
Table of Contents

The EM-DAT dataset catalogs over 26,000 mass disasters worldwide from 1900 to present day.
EM-DAT is maintained by the Centre for Research on the Epidemiology of Disasters (CRED), and is provided in open access to any person or organization performing non-commercial research.
It is critical to understand that EM-DAT does not catalog all natural disasters worldwide. It instead focuses on mass disasters.
According to EM-DAT and CRED, a mass disaster is a specific type of natural disaster that leads to significant human and economic loss, requiring that at least one of the following criteria hold:
For example, if a tornado touches down in rural Texas but doesn’t kill anyone, nor does it cause significant property damage, then the tornado would not be added to the EM-DAT dataset.
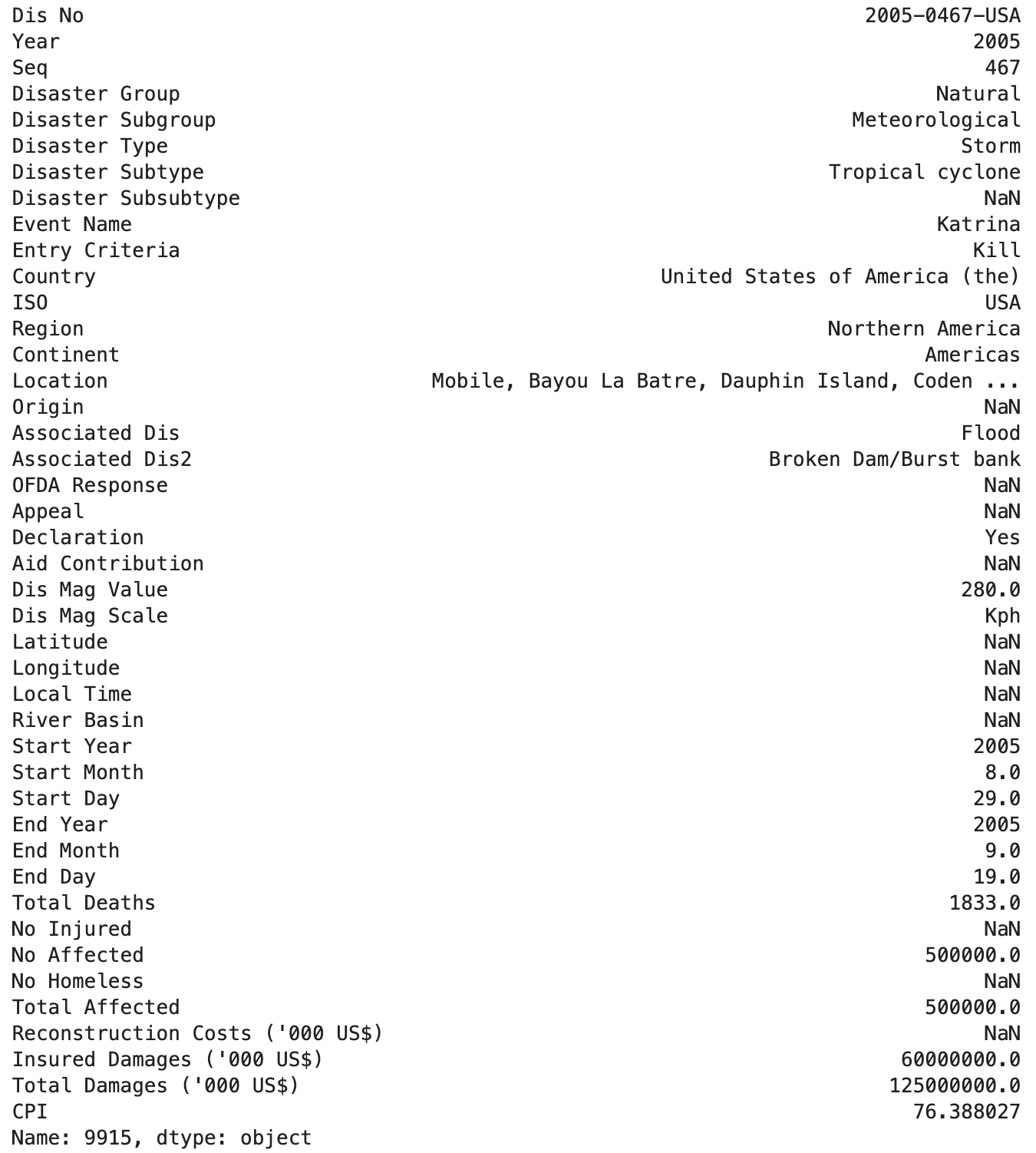
An example of a mass natural disaster that would be included in the the EM-DAT dataset is Hurricane Katrina, which absolutely devastated New Orleans, LA, and the surrounding areas, causing over 1,800 fatalities and estimated damages in the range of $98-145 billion USD.
Clearly, Hurricane Katrina passes multiple tests for inclusion in the EM-DAT dataset.
Below, I’ve included the data EM-DAT reports for Hurricane Katrina so you can get a feel for the data EM-DAT provides:
It’s also worth nothing that EM-DAT utilizes a hierarchical classification of all disasters, as I discuss in the “Breaking down disaster types in the EM-DAT dataset” section of the article.
One particular benefit of the hierarchical structure is that you can “drill down” into natural disaster type based on the following taxonomy:
Let’s take “extreme storms” as an example:
Using Python and Pandas, we can easily filter the natural disasters we are most interested in.
 Exploring the EM-DAT dataset (image credit)
Exploring the EM-DAT dataset (image credit)
Approximately two-thirds of disasters recorded in EM-DAT are related to natural hazards.
Here are common occurrences for natural hazards in the dataset:
As you can see, the EM-DAT dataset is quite diverse, covering a wide variety of natural disasters.
While there are certainly other datasets that provide far more data for a specific natural disaster type (for example, NOAA’s dataset on tornadoes), there are few datasets like EM-DAT that provides worldwide natural disaster data for such a diverse collection of disaster types.
The diversification factor alone makes EM-DAT worth studying in more detail (at least in my humble opinion).
 The EM-DAT dataset is not without its limitations (image credit)
The EM-DAT dataset is not without its limitations (image credit)
I want to be upfront and say that working with the EM-DAT dataset can be a bit challenging at times:
For example, when working with location data in the United States, some rows will have a mixture of:
Much of this location data is mixed and matched, making it sometimes challenging to work with.
Note: In fact, I had to develop a separate AI-based script to clean up the location data (which I’ll provide in a future article).
While EM-DAT strives to maintain high data accuracy, it relies on multiple sources for its data, including:
Each of these organizations has its own reporting standards which may or may not facilitate high data accuracy.
Additionally, the estimates on economic loss data can vary significantly, implying that the economic loss estimates EM-DAT reports may not measure the full extent of the natural disaster impact.
Finally, and in my opinion, most importantly, under-reporting can be a concern when utilizing EM-DAT in your work or research.
By design, EM-DAT only includes natural disasters where:
By definition, these exclusion criteria may result in natural disasters not being included in the dataset (but otherwise should be) due to limited media coverage, lack of money/infrastructure to gather reliable data, etc.
The EM-DAT is an invaluable resource, but it does have its limitations, so keep them in mind if you choose to utilize it in your own work;
The EM-DAT dataset includes 43 features (i.e., columns) used to represent and quantify mass natural disasters worldwide.
I’m still exploring and wrapping my head around the nuanced contextualization of each of these features, but I’ve done my best to summarize each of them below:
yes or NaN).yes, no, or NaN).yes, no, or NaN).110, and Dis Mag Scale reports KPH, then the natural disaster was reported as having a (presumed) wind speed of 110 KPH.Again, I want to reiterate that these explanations are provided at the best of my knowledge given my current understanding of the dataset.
If you are already an expert in the EM-DAT dataset (or better yet, a curator of the dataset), please leave a comment below to correct any of my unfortunate (but well intentioned) misgivings.
When confronted with a new dataset that I have no prior experience with, I first like to perform an Exploratory Data Analysis (EDA) to better understand and summarize its main characteristics.
“In statistics, exploratory data analysis is an approach of analyzing data sets to summarize their main characteristics, often using statistical graphics and other data visualization methods”
To start, I like to build a basic table that provides a summary of the following information:
The following code loads the EM-DAT dataset from disk:
# import the necessary packages
from collections import namedtuple
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
import os
# specify the path to the EM-DAT dataset
emdat_dataset_path = os.path.join(
"natural-disasters-data",
"em-dat",
"EMDAT_1900-2021_NatDis.csv"
)
# load the EM-DAT natural disasters dataset from disk
df = pd.read_csv(emdat_dataset_path)
df.head()
Note: For the sake of simplicity (and to ensure you can reproduce my results), I am using the version of the EM-DAT dataset hosted on Kaggle rather than the one provided by Centre for Research on the Epidemiology of Disasters (which requires registration).
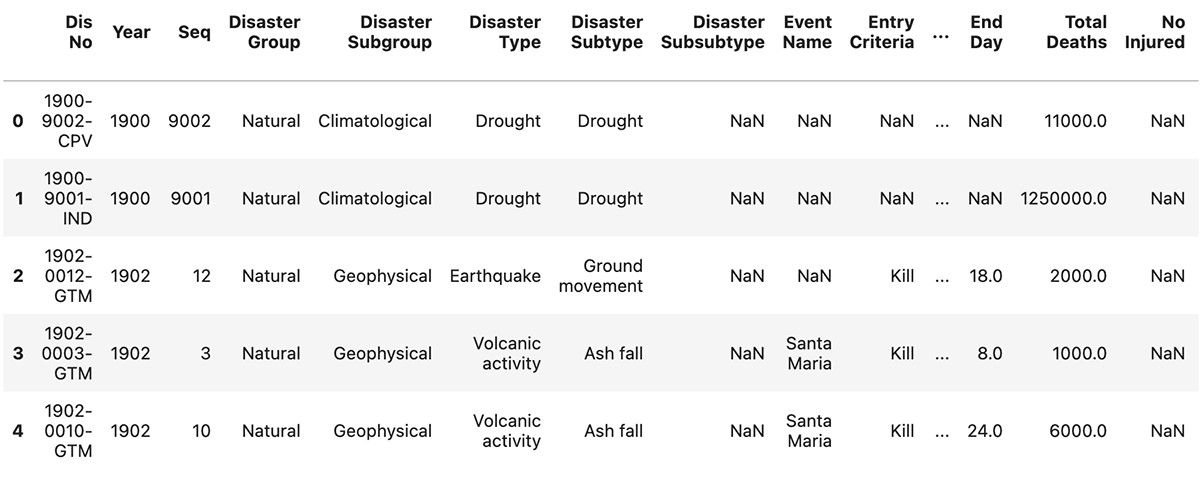
Below are the first five rows of the dataset:
| Dis No | Year | Seq | Disaster Group | Disaster Subgroup | Disaster Type | Disaster Subtype | Disaster Subsubtype | Event Name | Entry Criteria | Country | ISO | Region | Continent | Location | Origin | Associated Dis | Associated Dis2 | OFDA Response | Appeal | Declaration | Aid Contribution | Dis Mag Value | Dis Mag Scale | Latitude | Longitude | Local Time | River Basin | Start Year | Start Month | Start Day | End Year | End Month | End Day | Total Deaths | No Injured | No Affected | No Homeless | Total Affected | Reconstruction Costs ('000 US$) | Insured Damages ('000 US$) | Total Damages ('000 US$) | CPI | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1900-9002-CPV | 1900 | 9002 | Natural | Climatological | Drought | Drought | NaN | NaN | NaN | Cabo Verde | CPV | Western Africa | Africa | Countrywide | NaN | Famine | NaN | NaN | No | No | NaN | NaN | Km2 | NaN | NaN | NaN | NaN | 1900 | NaN | NaN | 1900 | NaN | NaN | 11000.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 3.261389 |
| 1 | 1900-9001-IND | 1900 | 9001 | Natural | Climatological | Drought | Drought | NaN | NaN | NaN | India | IND | Southern Asia | Asia | Bengal | NaN | NaN | NaN | NaN | No | No | NaN | NaN | Km2 | NaN | NaN | NaN | NaN | 1900 | NaN | NaN | 1900 | NaN | NaN | 1250000.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 3.261389 |
| 2 | 1902-0012-GTM | 1902 | 12 | Natural | Geophysical | Earthquake | Ground movement | NaN | NaN | Kill | Guatemala | GTM | Central America | Americas | Quezaltenango, San Marcos | NaN | Tsunami/Tidal wave | NaN | NaN | NaN | NaN | NaN | 8.0 | Richter | 14 | -91 | 20:20 | NaN | 1902 | 4.0 | 18.0 | 1902 | 4.0 | 18.0 | 2000.0 | NaN | NaN | NaN | NaN | NaN | NaN | 25000.0 | 3.391845 |
| 3 | 1902-0003-GTM | 1902 | 3 | Natural | Geophysical | Volcanic activity | Ash fall | NaN | Santa Maria | Kill | Guatemala | GTM | Central America | Americas | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 1902 | 4.0 | 8.0 | 1902 | 4.0 | 8.0 | 1000.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 3.391845 |
| 4 | 1902-0010-GTM | 1902 | 10 | Natural | Geophysical | Volcanic activity | Ash fall | NaN | Santa Maria | Kill | Guatemala | GTM | Central America | Americas | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 1902 | 10.0 | 24.0 | 1902 | 10.0 | 24.0 | 6000.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 3.391845 |
A few noteworthy items:
Below is my basic exploratory data analysis function (which I optimized from Juliana’s original implementation):
def basic_eda(df):
# count the number of duplicated rows, then grab all NaN (i.e., null) rows
# in the dataframe
num_duplicated = df.duplicated().sum()
is_nan = df.isnull()
# count the total number of rows that contain *at least one* null value
num_null_rows = is_nan.any(axis=1).sum()
# count the total number of null values across *all* rows and *all* columns
# (i.e., a sum of a sum)
num_total_null = df.isnull().sum().sum()
# construct a named tuple to represent each row in the exploratory data
# analysis summary
EDARow = namedtuple("EDARow", ["Name", "Value", "Notes"])
# build the list of exploratory data analysis rows
rows = [
EDARow("Samples", df.shape[0], ""),
EDARow("Features", df.shape[1], ""),
EDARow("Duplicate Rows", num_duplicated, ""),
EDARow("Rows with NaN", num_null_rows, "{:.2f}% all rows".format(
(num_null_rows / df.shape[0]) * 100)),
EDARow("Total NaNs", num_total_null, "{:.2f}% feature matrix".format(
(num_total_null / (df.shape[0] * df.shape[1])) * 100)),
]
# build and return our exploratory data analysis dataframe
return pd.DataFrame(rows, columns=["Name", "Value", "Notes"])
The above code builds a summary dataframe.
An aspect of the code I want to point out is the difference between:
num_null_rows)num_total_null)The former is accomplished simply by summing the number of rows that have at least one null column.
I accomplish the later by doing two sums:
The final dataframe provides the following information:
I can then call the basic_eda method on the EM-DAT dataset:
# perform a basic exploratory data analysis of the EM-DAT dataset
basic_eda(df)
Which provides the following summary table:
| Name | Value | Notes | |
|---|---|---|---|
| 0 | Samples | 15827 | |
| 1 | Features | 43 | |
| 2 | Duplicate Rows | 0 | |
| 3 | Rows with NaN | 15827 | 100.00% all rows |
| 4 | Total NaNs | 285923 | 42.01% feature matrix |
As the EDA table demonstrates, there are a total of 15,827 rows in the EM-DAT dataset.
Note: Again, I want to stress that I’m using the Kaggle version of the EM-DAT dataset, which is easily accessible and ensures you can reproduce any of my results. You can also use the CRED version of EM-DAT, which is also official version, but will require you to register before you can access it.
There are 43 features (i.e., columns) in the EM-DAT dataset (each of which I did my best to summarize earlier in this article).
The EM-DAT dataset has zero duplicate rows, meaning that no two rows in the dataset have identical column values.
Earlier in this article I mentioned that the EM-DAT natural disaster dataset is noisy, containing missing and incomplete data — the final two rows in the EDA table provide evidence to this claim.
First, every single row in the dataset has at least one column with a NaN value, meaning that there is no row with all column/feature values provided.
Additionally, approximately 42% of the entire feature matrix (15827 x 43 = 68,0561) is also NaN, implying that nearly half the dataset contains null values.
Datasets with a large number of NaNs can be quite challenging and problematic to work with for data scientists as we need to define how to handle missing values:
In many cases, the answer is dataset (and even feature/column) dependent.
I’ve found that some of EM-DAT features can be reliably filled in while others cannot. I’ll likely do a future article on this type of study.
Given that 42% of all values in EM-DAT are missing, I found it worthwhile to spend a bit more time investigating exactly which features have values provided, and which ones are predominantly NaN.
Furthermore, I wanted to understand the uniqueness of the features, as unique values can have predictive power in downstream machine learning models.
To that end, I again built on Juliana’s work, and defined the summarize_data function to further summarize the EM-DAT dataset:
def summarize_data(df):
# initialize a summary dataframe consiting of the original dataframe's
# column names and data types
summary = pd.DataFrame(df.dtypes, columns=["dtypes"])
# reset the summary index, rename the "index" column to "Name", and then
# remove the "index" column
summary = summary.reset_index()
summary["Name"] = summary["index"]
summary = summary[["Name", "dtypes"]]
# count the number of (1) null values for each column, and (2) the unique
# values in each column
summary["Missing"] = df.isnull().sum().values
summary["Uniques"] = df.nunique().values
# return the summary dataframe
return summary
The summarize_data method constructs a dataframe that analyzes every column of the dataset.
For each column, four values are provided:
Calling summarize_data on the df is as simple as:
# summarize the EM-DAT dataframe
summarize_data(df)
Which provides the following large block of output, analyzing the total number of missing and unique values for each column:
| Name | dtypes | Missing | Uniques | |
|---|---|---|---|---|
| 0 | Dis No | object | 0 | 15827 |
| 1 | Year | int64 | 0 | 122 |
| 2 | Seq | int64 | 0 | 1266 |
| 3 | Disaster Group | object | 0 | 1 |
| 4 | Disaster Subgroup | object | 0 | 6 |
| 5 | Disaster Type | object | 0 | 15 |
| 6 | Disaster Subtype | object | 2984 | 27 |
| 7 | Disaster Subsubtype | object | 14782 | 12 |
| 8 | Event Name | object | 12024 | 1532 |
| 9 | Entry Criteria | object | 335 | 13 |
| 10 | Country | object | 0 | 227 |
| 11 | ISO | object | 0 | 227 |
| 12 | Region | object | 0 | 23 |
| 13 | Continent | object | 0 | 5 |
| 14 | Location | object | 1808 | 12453 |
| 15 | Origin | object | 12190 | 638 |
| 16 | Associated Dis | object | 12614 | 30 |
| 17 | Associated Dis2 | object | 15145 | 30 |
| 18 | OFDA Response | object | 14220 | 1 |
| 19 | Appeal | object | 13259 | 2 |
| 20 | Declaration | object | 12612 | 2 |
| 21 | Aid Contribution | float64 | 15150 | 556 |
| 22 | Dis Mag Value | float64 | 10926 | 1859 |
| 23 | Dis Mag Scale | object | 1171 | 5 |
| 24 | Latitude | object | 13111 | 2360 |
| 25 | Longitude | object | 13108 | 2426 |
| 26 | Local Time | object | 14735 | 777 |
| 27 | River Basin | object | 14570 | 1184 |
| 28 | Start Year | int64 | 0 | 122 |
| 29 | Start Month | float64 | 384 | 12 |
| 30 | Start Day | float64 | 3600 | 31 |
| 31 | End Year | int64 | 0 | 122 |
| 32 | End Month | float64 | 708 | 12 |
| 33 | End Day | float64 | 3532 | 31 |
| 34 | Total Deaths | float64 | 4591 | 840 |
| 35 | No Injured | float64 | 12011 | 753 |
| 36 | No Affected | float64 | 6844 | 3486 |
| 37 | No Homeless | float64 | 13422 | 906 |
| 38 | Total Affected | float64 | 4471 | 4982 |
| 39 | Reconstruction Costs ('000 US$) | float64 | 15796 | 29 |
| 40 | Insured Damages ('000 US$) | float64 | 14735 | 359 |
| 41 | Total Damages ('000 US$) | float64 | 10661 | 1558 |
| 42 | CPI | float64 | 424 | 111 |
However, I find looking at such a table tedious and uninformative.
A more useful exercise is to visualize the above information.
In my opinion, a better way to explore features with missing values is to count the number of NaN values per column, and then create a bar chart that plots the counts in descending order.
Doing so makes it trivially easy to identify columns with a large number of missing values with a simple visual inspection.
The following plot_null_columns method is more verbose, but provides a nicer, cleaner analysis:
def plot_null_columns(
df,
title,
x_label="Feature Names",
y_label="# of Null Values",
figsize=(20, 5)
):
# count the number of times a given column has a null value
null_cols = df.isnull().sum().sort_values(ascending=False)
# initialize the figure, set the tick information, and update the spines
plt.figure(figsize=figsize)
sns.set(style="ticks", font_scale=1)
plt.xticks(rotation=90, fontsize=12)
sns.despine(top=True, right=True, bottom=False, left=True)
# plot the data
ax = sns.barplot(x=null_cols.index, y=null_cols, palette="cool_r")
# set the x-label, y-label, and title
ax.set_xlabel(x_label)
ax.set_ylabel(y_label)
plt.title(title)
# loop over the patches and null column counts
for (p, count) in zip(ax.patches, null_cols):
# compute the percentage of the number of rows that have a null value
# for the current column
ax.annotate(
"{:.1f}%".format((count / df.shape[0]) * 100),
(p.get_x() + (p.get_width() / 2.0), abs(p.get_height())),
ha="center",
va="bottom",
rotation="vertical",
color="black",
xytext=(0, 10),
textcoords="offset points"
)
What I found tricky about the above code block is figuring out the computation of null_cols such that it could also be provided to the barplot method to plot the number of columns with missing values in descending order.
The trick was to first compute a column-wise count of NaNs, sort them in descending order (via sort_values), and then provide null_cols.index to the barplot function.
Secondly, I find it extremely helpful to also plot the percentage of null counts for each column directly above each of the individual bars — the for loop handles that computation.
The handy plot_null_columns function can then be called via:
# plot the null column counts within the dataset
plot_null_columns(df, "Features With Missing Values")
Which produces the following plot:
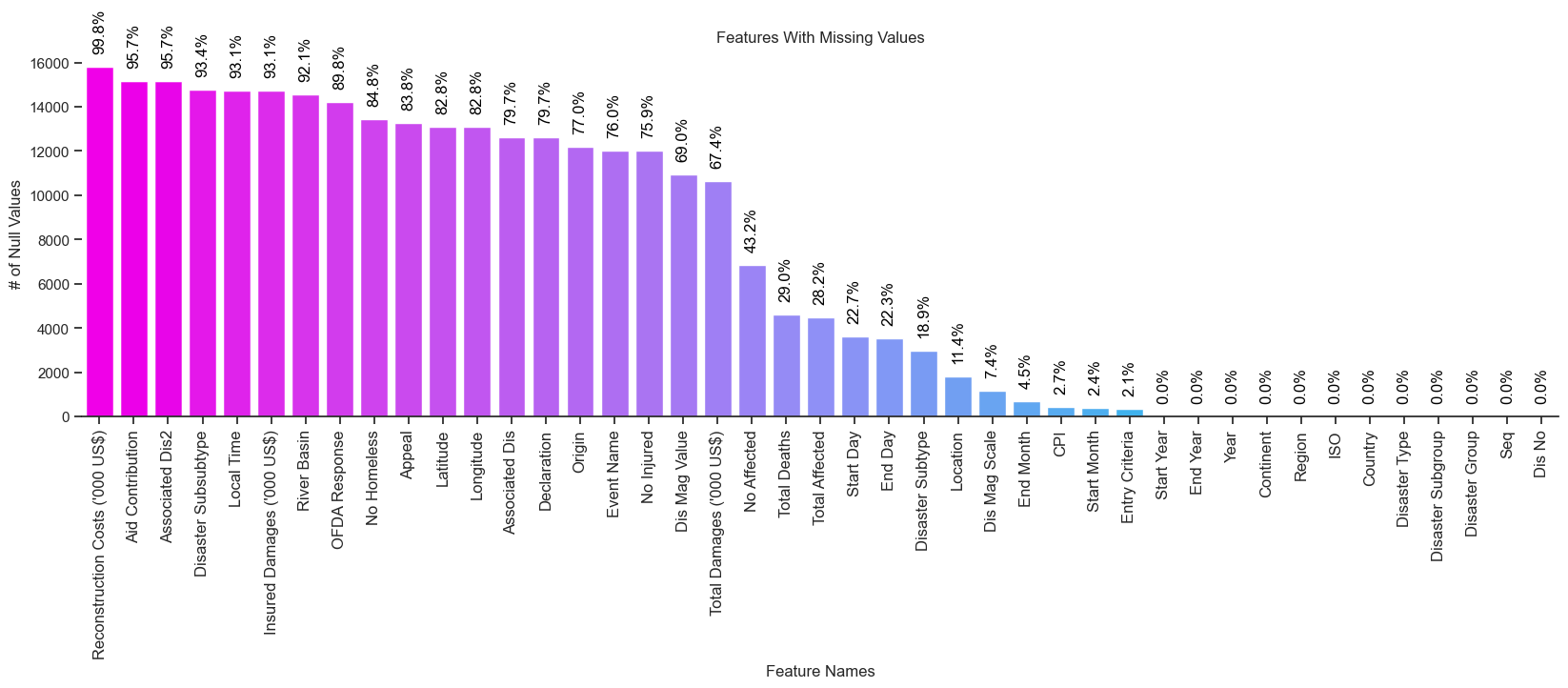
Clearly, it’s far easier to visually identify features with missing values using this approach.
A quick inspection of the above plot reveals there are seven features where over 90% of values are missing for a particular column, including:
Conversely, there are twelve features that I would consider form the “core definition” of the EM-DAT dataset:
All twelve of these features contain values (i.e., non are missing/NaN), and while these values may still be noisy, they are at least present in the EM-DAT dataset.
In my initial studies I’ve predominately focused on these core features.
My initial foray into the EM-DAT dataset concluded with an exploration of the natural disaster hierarchy grouping using Disaster Group as the base, and then becoming more fine-grained based on:
I found the hierarchy exploration absolutely essential in understanding EM-DAT’s structure, and if you’re exploring EM-DAT for the first time, I’d suggest you spend considerable time here.
Below is the code I used:
# define the disaster type columns we are interested then
disaster_cols = [
"Disaster Subgroup",
"Disaster Type",
"Disaster Subtype",
"Disaster Subsubtype",
]
# grab the disaster data from the dataframe
disaster_df = df[disaster_cols]
# fill any null values with an empty string (implying that no subgroups or
# subtypes exist for the current value)
disaster_df = disaster_df.fillna(value={
"Disaster Subtype": "NA",
"Disaster Subsubtype": "NA",
})
# construct the final dataframe which displays a hierarchical overview of the
# disaster types, including the counts for each one
disaster_df = pd.DataFrame(
disaster_df.groupby(disaster_cols).size().to_frame("count")
)
disaster_df
Basically, what I’m doing here is:
disaster_cols from the dataframeThe groupby function call is doing quite a bit, but the gist is that the code groups the dataframe by the unique combinations of disaster_cols, counting the occurrences of each combination. The final counts for each combination are stored in a new column named count.
Running the above code produces the following table which concisely depicts the hierarchical organization of natural disasters in EM-DAT:
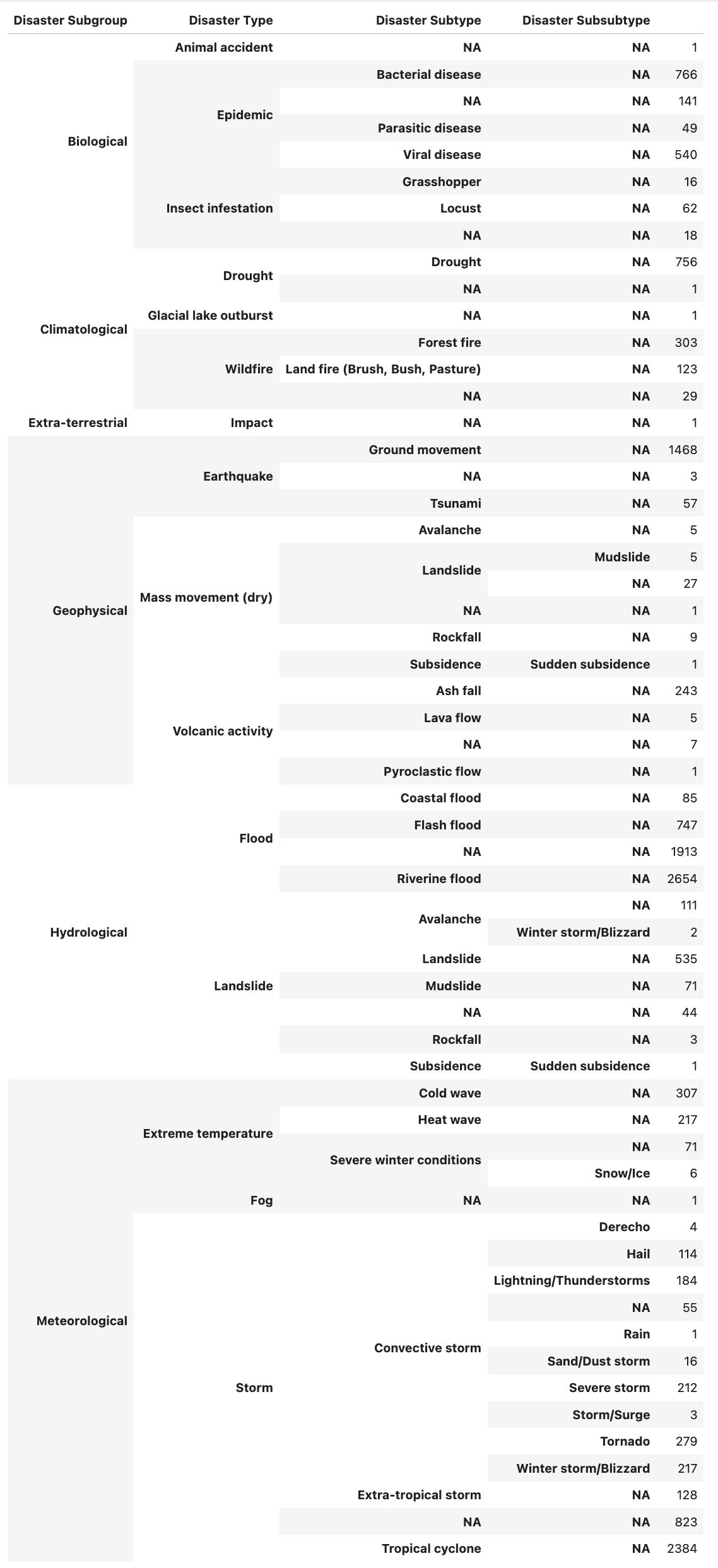
For example, let’s investigate the “Biological” subgroup — there are three disaster types for this subgroup:
Now, let’s further examine the “Insect infestation” disaster type:
I had to sit and study this hierarchy table for a bit to fully appreciate and understand the EM-DAT disaster categorization hierarchy.
If you’re considering working with EM-DAT, I’d highly suggest you spend at least 30-60 minutes here before you move on to more detailed data analysis.
If I hadn’t taken the time to understand the data hierarchy, I think I would have struggled considerably when I moved on to more advanced analysis.
Adrian Rosebrock. “Getting Started with the International Disaster Database (EM-DAT) with Python and Pandas”, NaturalDisasters.ai, 2023, https://naturaldisasters.ai/posts/getting-started-em-dat-international-disaster-database/.
@incollection{ARosebrock_GettingStartedEMDAT,
author = {Adrian Rosebrock},
title = {Getting Started with the International Disaster Database (EM-DAT) using Python and Pandas},
booktitle = {NaturalDisasters.ai},
year = {2023},
url = {https://naturaldisasters.ai/posts/getting-started-em-dat-international-disaster-database/},
}
AI generated content disclaimer: I’ve used a sprinkling of AI magic in this blog post, namely in the following sections:
Don’t fret, my human eyeballs have read and edited every word of the AI generated content, so rest assured, what you’re reading is as accurate as I possibly can make it. If there are any discrepancies or inaccuracies in the post, it’s my fault, not that of our machine assistants.